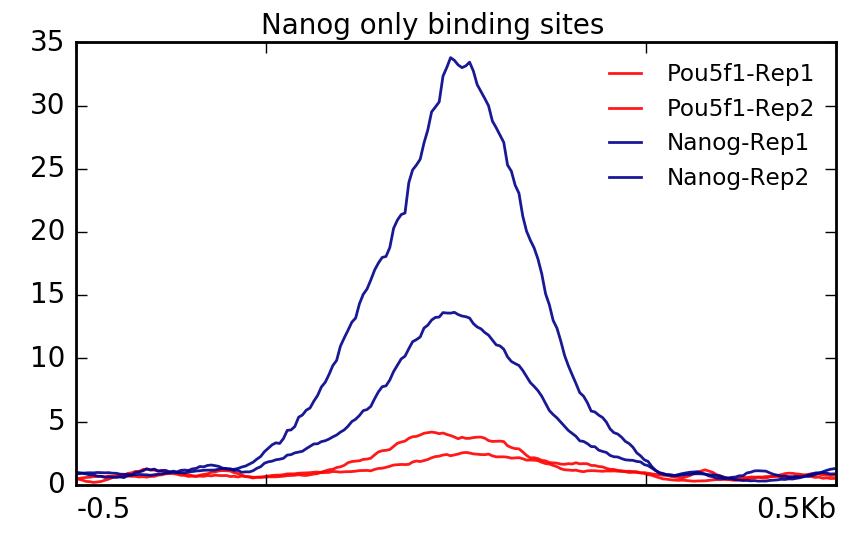
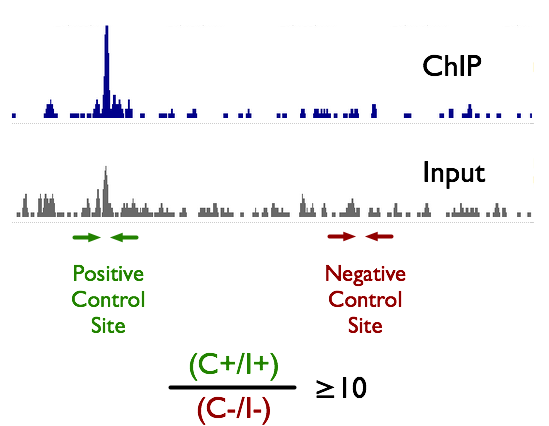
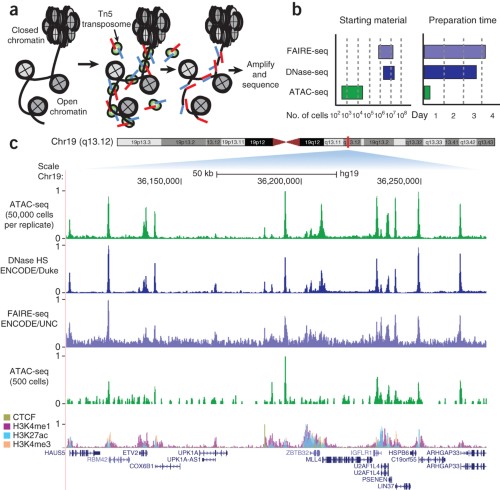
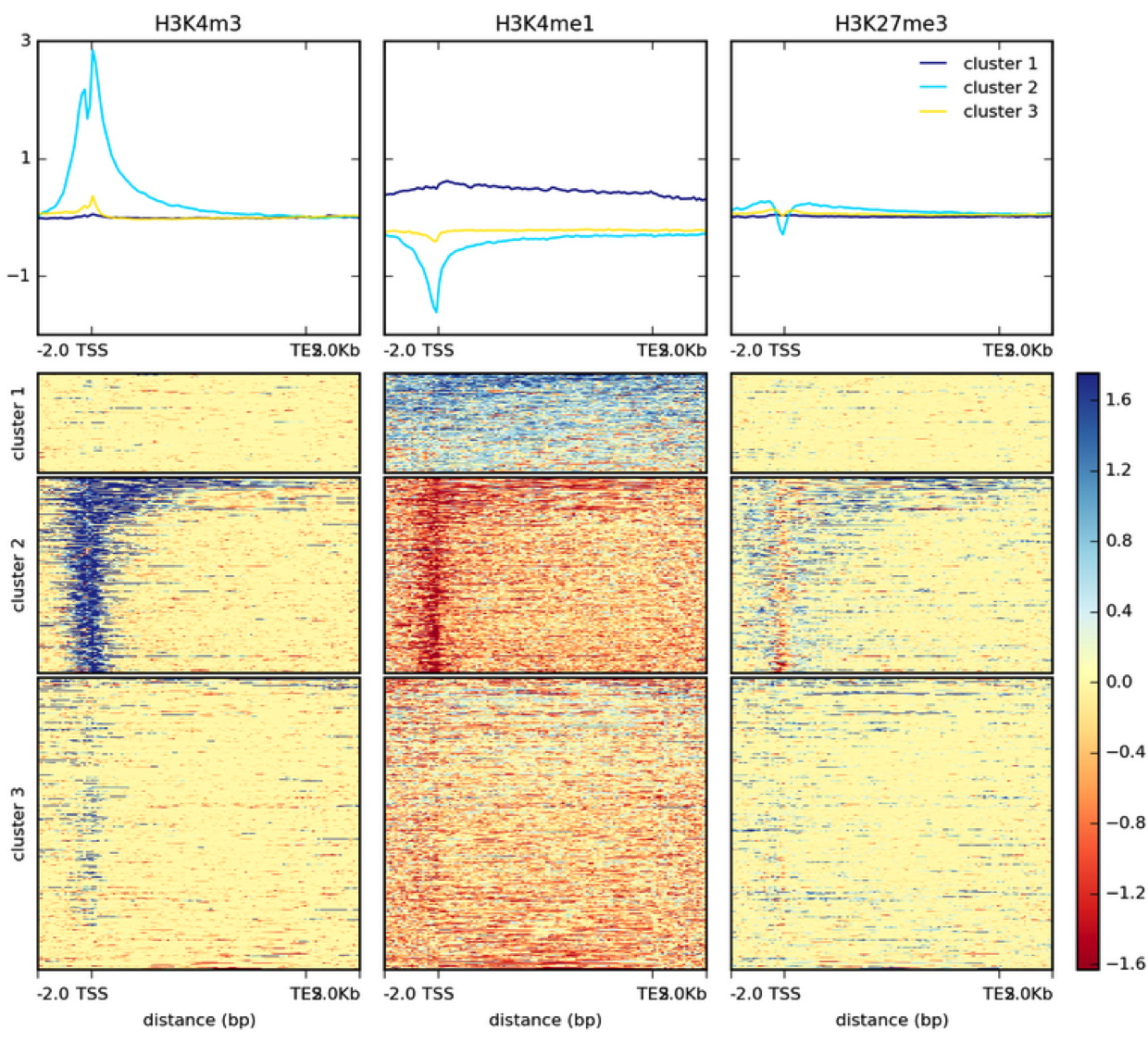
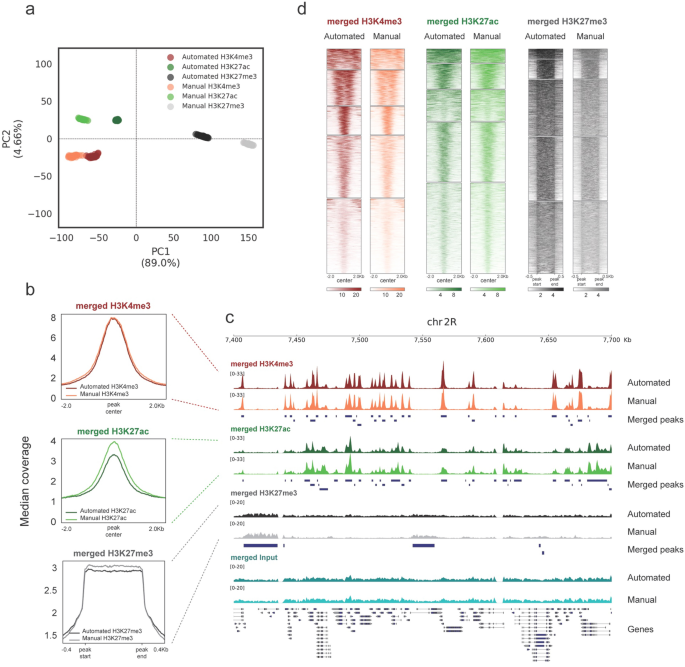
TAF-ChIP: an ultra-low input approach for genome-wide chromatin immunoprecipitation assay | Life Science Alliance
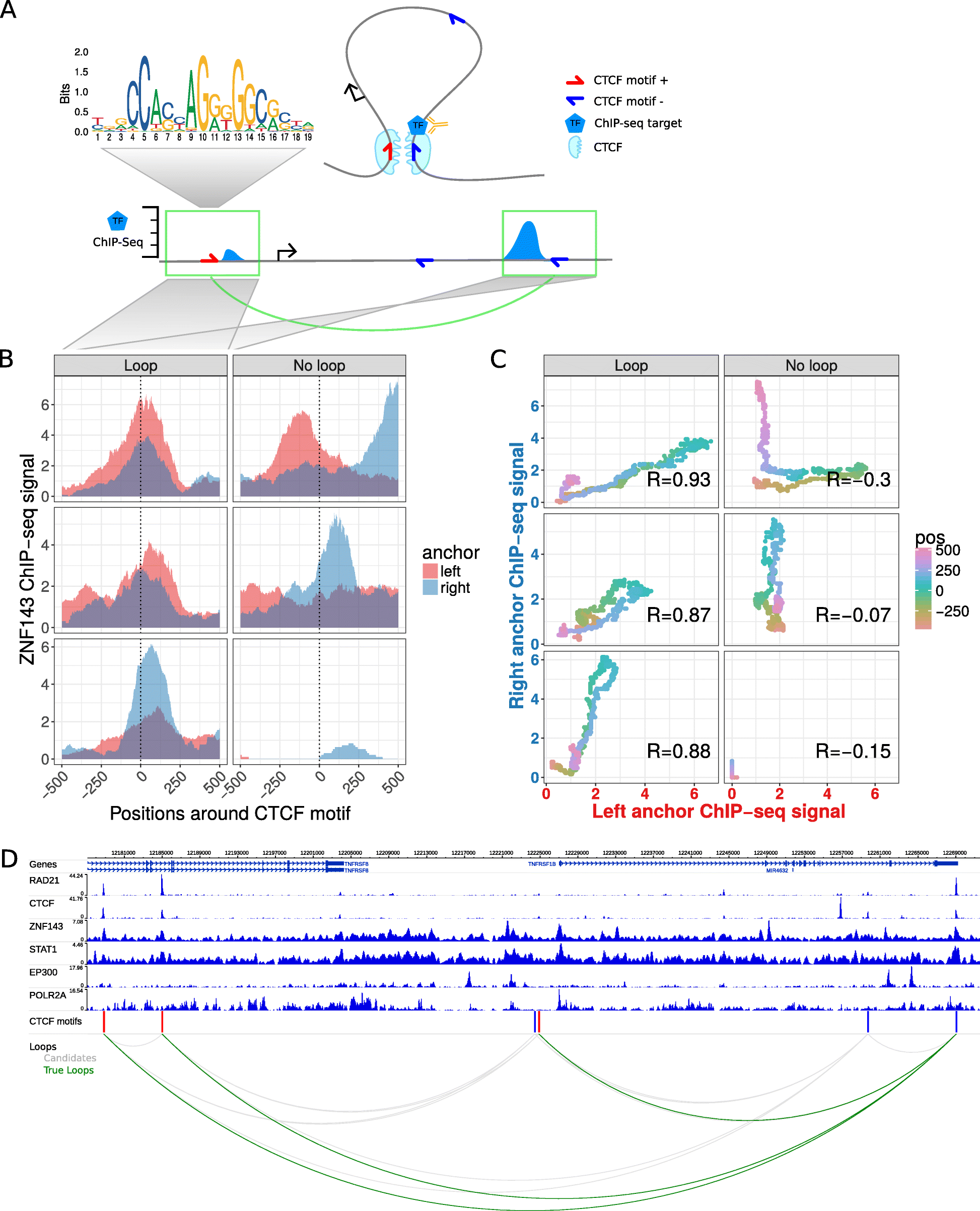
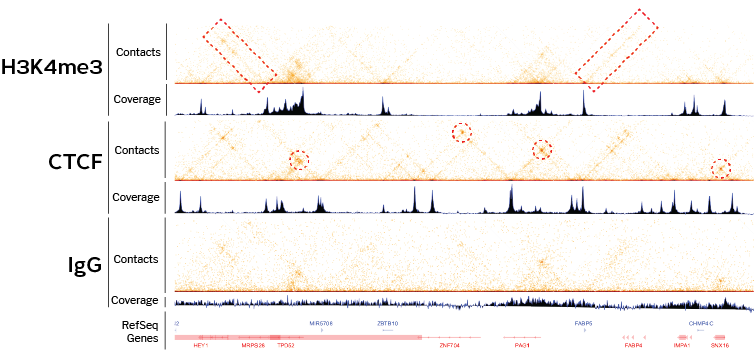
7C: Computational Chromosome Conformation Capture by Correlation of ChIP-seq at CTCF motifs | BMC Genomics | Full Text

Example genome browser screen-shot. ChIP-seq read coverage of H3K27me3... | Download Scientific Diagram

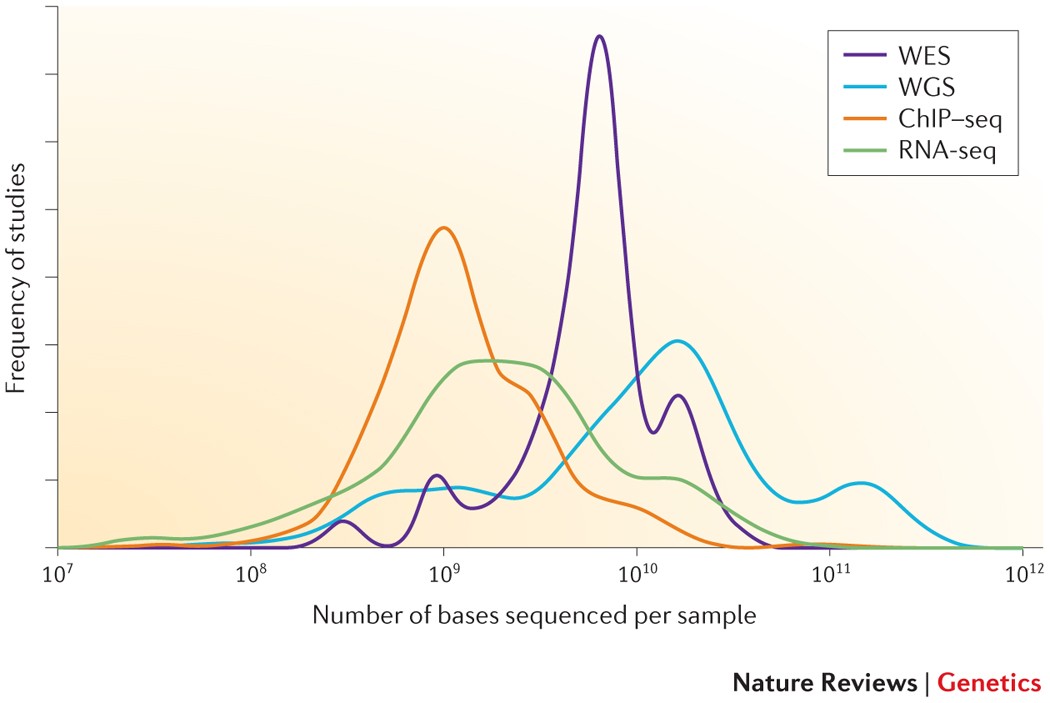
Seq-ing answers: Current data integration approaches to uncover mechanisms of transcriptional regulation - ScienceDirect

Vezf1 protein binding sites genome-wide are associated with pausing of elongating RNA polymerase II | PNAS

Clustering Genes Based on Patterns of Pol II ChIP-Seq Coverage around... | Download Scientific Diagram
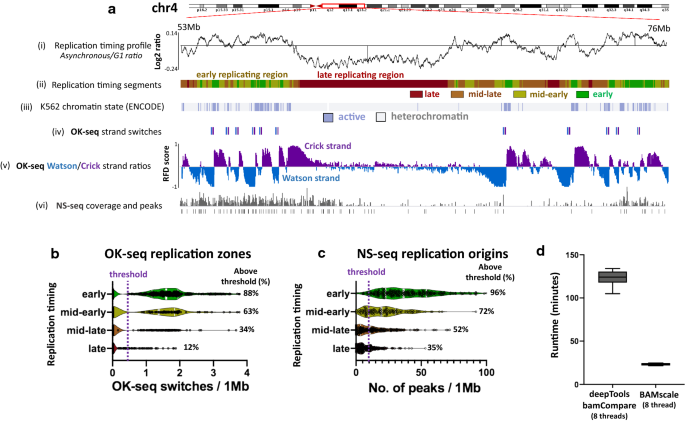
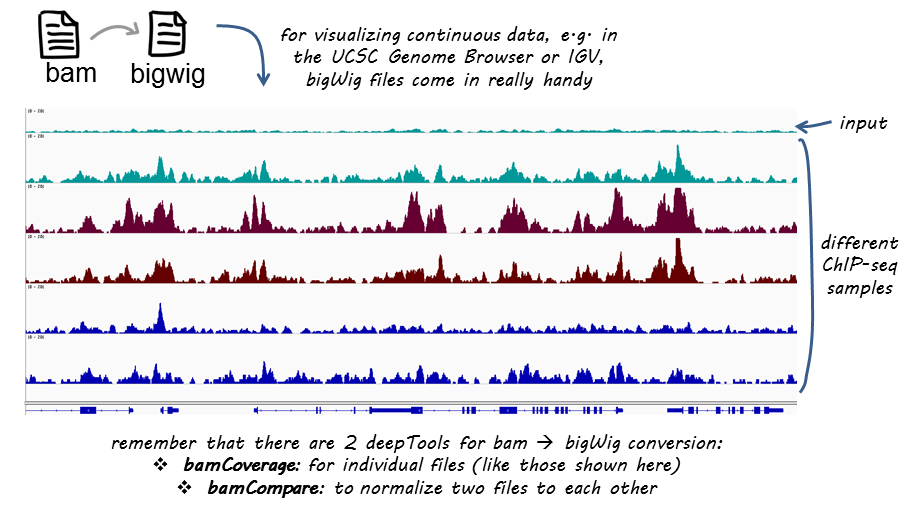
BAMscale: quantification of next-generation sequencing peaks and generation of scaled coverage tracks | Epigenetics & Chromatin | Full Text

Detecting, Categorizing, and Correcting Coverage Anomalies of RNA-Seq Quantification - ScienceDirect